Automated Assessment of Corneal Nerves using Deep Learning
Automated Assessment of Corneal Nerves using Deep Learning
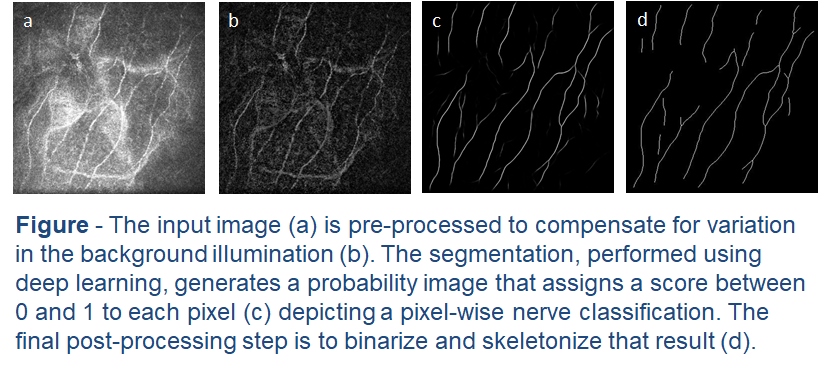
In vivo corneal microscopy has seen a recent resurgence in interest among both the research and clinical communities. The cornea is the most densely innervated tissue in the human body so the ability to capture near cellular resolution images of its different layers using an office-based device offers significant and largely untapped clinical utility. Using image processing, it is now possible to automatically and accurately trace the nerve fibers seen in the sub-basal plexus of the cornea. This analysis generates a number of compelling biomarkers that have utility in the management of a variety of diseases including diabetes, Alzheimer’s, Parkinson’s, HIV and SIV. This interest has led to Heidelberg Engineering bringing back their HRT3 imaging device specifically for corneal imaging and promises to realize the full potential of this modality.
Professor Mankowski’s lab at Johns Hopkins University School of Medicine has been investigating animal models of peripheral neuropathy for a number of years. Since 2013, Voxeleron has been working with them on image processing for the automatic detection and quantification of corneal sub-basal nerves. We reported our initial findings in [Dorsey 2014] and [Misra 2017] in both animal and clinical data. More recent work, using deep convolutional neural networks, was first given in [Oakley 2018] and now [Oakley 2020], the subject of this post.
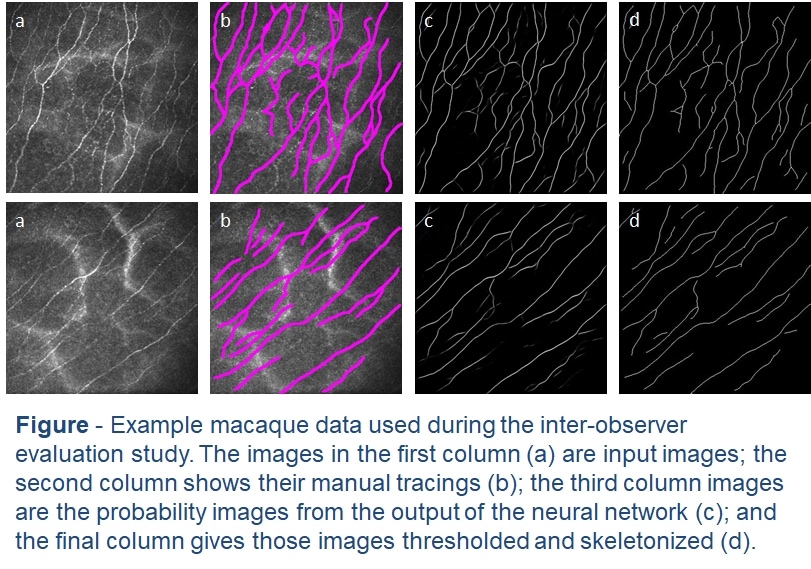
 This latest work assesses the accuracy of different deep-learning models on an early macaque data set. In the tradition of early deep learning articles, we entitled the best performing model DeepNerve. A sequestered data set from a similar macaque population was then used to validate the performance of DeepNerve and also performed an inter-observer study. Performance was shown to be highly accurate, and the automated method could not be told apart from the three expert readers. Based on this validation effort, we are now using the algorithm in animal studies [McCarron 2020].
This latest work assesses the accuracy of different deep-learning models on an early macaque data set. In the tradition of early deep learning articles, we entitled the best performing model DeepNerve. A sequestered data set from a similar macaque population was then used to validate the performance of DeepNerve and also performed an inter-observer study. Performance was shown to be highly accurate, and the automated method could not be told apart from the three expert readers. Based on this validation effort, we are now using the algorithm in animal studies [McCarron 2020].
Please contact deepnerve@voxeleron.com for additional information.
References
[Dorsey 2014] – “Loss of Corneal Sensory Nerve Fibers in SIV-infected Macaques: An Alternate Approach to Investigate HIV-induced PNS Damage,” by Jamie L. Dorsey, Lisa M. Mangus, Jonathan D. Oakley, et al. The American Journal of Pathology, Volume 184, Issue 6 (June 2014) published by Elsevier.http://ajp.amjpathol.org/article/S0002-9440(14)00162-X/fulltext
[Misra 2017] – “Automated analysis of in vivo confocal microscopy images of corneal nerves”, Stuti L Misra, Jonathan D Oakley, Charles N McGhee, 1 Ellen F Wang, Dipika V Patel, Patrick M Tarwater, Joseph L Mankowski. ARVO Meeting Abstracts, 2017.https://iovs.arvojournals.org/article.aspx?articleid=2637559&resultClick=1
[Oakley 2018] – “Automated Analysis of In Vivo Confocal Microscopy Corneal Images Using Deep Learning”, Jonathan Oakley, Daniel Russakoff, Rachel Weinberg, Megan McCarron, Samuel Brill, Stuti Misra, Charles McGhee, Joseph Mankowski. ARVO Meeting Abstracts, 2018.https://iovs.arvojournals.org/article.aspx?articleid=2694279&resultClick=1
[Oakley 2020] – “Deep Learning-Based Analysis of Macaque Corneal Sub-Basal Nerve Fibers in Confocal Microscopy Images”, Jonathan D. Oakley, Daniel B. Russakoff, Megan E. McCarron, Rachel L. Weinberg, Jessica M. Izzi, Stuti L. Misra, Charles N. McGhee, Joseph L. Mankowski. Eye and Vision (2020) 7:27 https://doi.org/10.1186/s40662-020-00192-5.https://eandv.biomedcentral.com/articles/10.1186/s40662-020-00192-5
[McCarron 2020] – “Combining In Vivo Corneal Confocal Microscopy with Deep Learning-based Analysis Reveals Sensory Nerve Fiber Loss in Acute SIV Infection”, ME McCarron, RL Weinberg, JM Izzi, SE Queen, SL Misra, DB Russakoff, J Oakley, J Mankowski. bioRxiv doi: https://doi.org/10.1101/2020.04.19.048926https://doi.org/10.1101/2020.04.19.048926